20-Minute Sepsis Inflammation Assessment
A rapid, quantitative host-response profile (IL-6, IL-10, CRP, PCT, TNF-α, lactate) that supports earlier recognition, risk stratification, and rule-in/rule-out decisions—before organ dysfunction begins.

Sepsis is one of the costliest and deadliest emergencies worldwide, yet clinicians are forced to treat it with incomplete data. While life-saving decisions must be made within the first 1–2 hours, today’s tools return results in 24–72 hours, leaving hospitals to rely on broad-spectrum antibiotics, delayed escalation, and trial-and-error management.
The core bottleneck is phenotyping—determining which antibiotics will actually work. Meanwhile, sepsis is not a single problem but a dynamic interplay between host inflammation, pathogens, and drug response. No existing platform measures all three domains together, leaving clinicians without real-time clarity on infection trajectory.
Guanine closes this gap with an integrated sepsis platform built for rapid, actionable clarity. It unifies host-response profiling, culture-free pathogen ID, and true antimicrobial phenotyping—directly from whole blood, without waiting for culture. Together, these capabilities allow ED teams to treat earlier, ICU teams to escalate with precision, and stewardship teams to finally act on comprehensive real-time data.
Guanine provides a structured, time-aligned diagnostic workflow that delivers host-response assessment, pathogen and resistance detection, and antimicrobial phenotyping within the early clinical decision window for suspected sepsis.
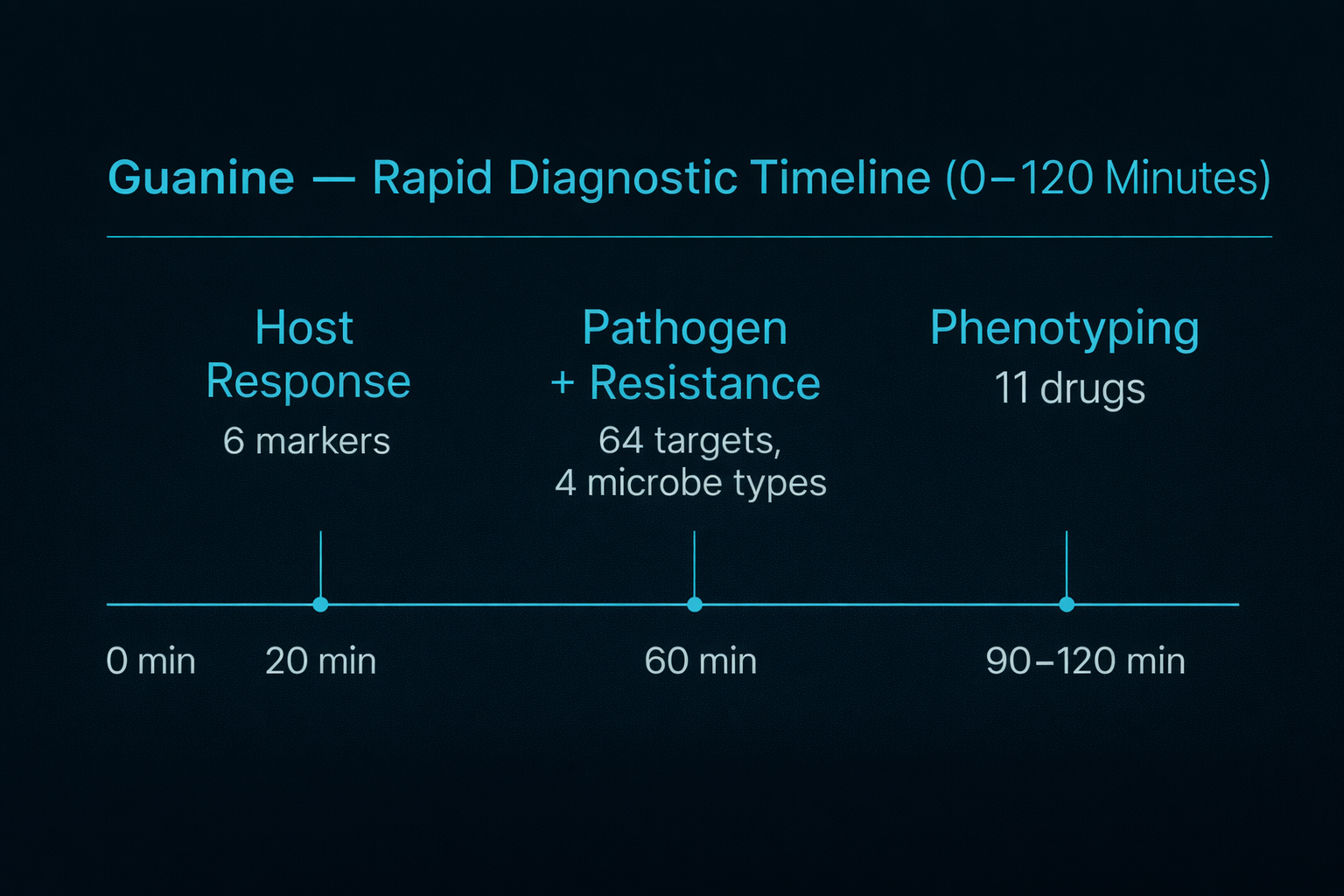
Initial host-response markers are quantified at approximately 20 minutes to support early risk stratification, followed by culture-free identification of pathogens and resistance targets at approximately 60 minutes. Real-time antimicrobial phenotyping is completed within 90–120 minutes, enabling earlier evidence-based optimization of antimicrobial therapy compared with conventional culture-dependent workflows.
A rapid, quantitative host-response profile (IL-6, IL-10, CRP, PCT, TNF-α, lactate) that supports earlier recognition, risk stratification, and rule-in/rule-out decisions—before organ dysfunction begins.
Culture-free detection of up to 64 bloodborne pathogens, including:
Plus an expanded panel of resistance genes, all from the same sample.
Electrochemical phenotyping that captures real-time drug-response kinetics across 11 priority antibiotics—providing actionable susceptibility data in under two hours instead of days.